Kernel Methods¶
- step 1: use a special type of mapping functions
- still map feature space to a higher dimensional space
- but computation of \(\phi(x) \cdot \phi(z)\) is easy (remember the multiplication of features in SVM optimization)
- step 2: rewrite the model s.t.
- the mapping \(\phi(x)\) never needs to be explicitly computed - i.e. never compute terms like \(w \cdot \phi(x) + b\)
- only work with \(\phi(x) \cdot \phi(z)\) - we call this the kernel function
Ex.
We can compute \(\phi(x) \cdot \phi(z)\) easily:
We call this a kernel function
What about the quadratic feature mapping?
Kernel in SVM¶
Rewriting the primal form of the SVM to use the mapped space doesn’t seem easy.
But we can rewrite the dual form easily!
Kernelized SVM
Now SVM is computing a linear boundary in the higher-dimensional space without actually transforming vectors
Predictions:
Formal Definition¶
Let’s use the example kernel \(K(x, z) = (x \cdot z)^2\) for \(\phi(x) = <x_1^2, \sqrt{2}x_1x_2, x_2^2>\)
- a kernel function is implicitly associated with some \(\phi\)
- \(\phi\) maps input \(x\in X\) to a higher dimensional space \(F\):
- \(\phi: X \to F\)
- kernel takes 2 inputs from \(X\) and outputs their similarity in F
- \(K: X \times X \to R\)
- once you have a kernel, you don’t need to know \(\phi\)
Mercer’s Condition
- a function can be a kernel function K if a suitable \(\phi\) exists
- \(\exists \phi\) s.t. \(K(x, z) = \phi(x) \cdot \phi(z)\)
- mathematically: K should be positive semi-definite; i.e. for all square integrable f s.t. \(\int f(x)^2 dx < \infty\)
- \(\int \int f(x)K(x, z)f(z)dxdz > 0\)
- sufficient and necessary to identify a kernel function
Constructing Kernels
We already know some proven basic kernel functions - given Mercer’s condition, we can construct new kernels!
- \(k(x, z) = k_1(x, z) + k_2(x, z)\) (direct sum)
- \(k(x, z) = \alpha k_1(x, z)\) (\(\forall \alpha > 0\) - scalar product)
Note
Example: given that k1 and k2 are kernels, prove \(k(x, z) = k_1(x, z) + k_2(x, z)\) is a kernel
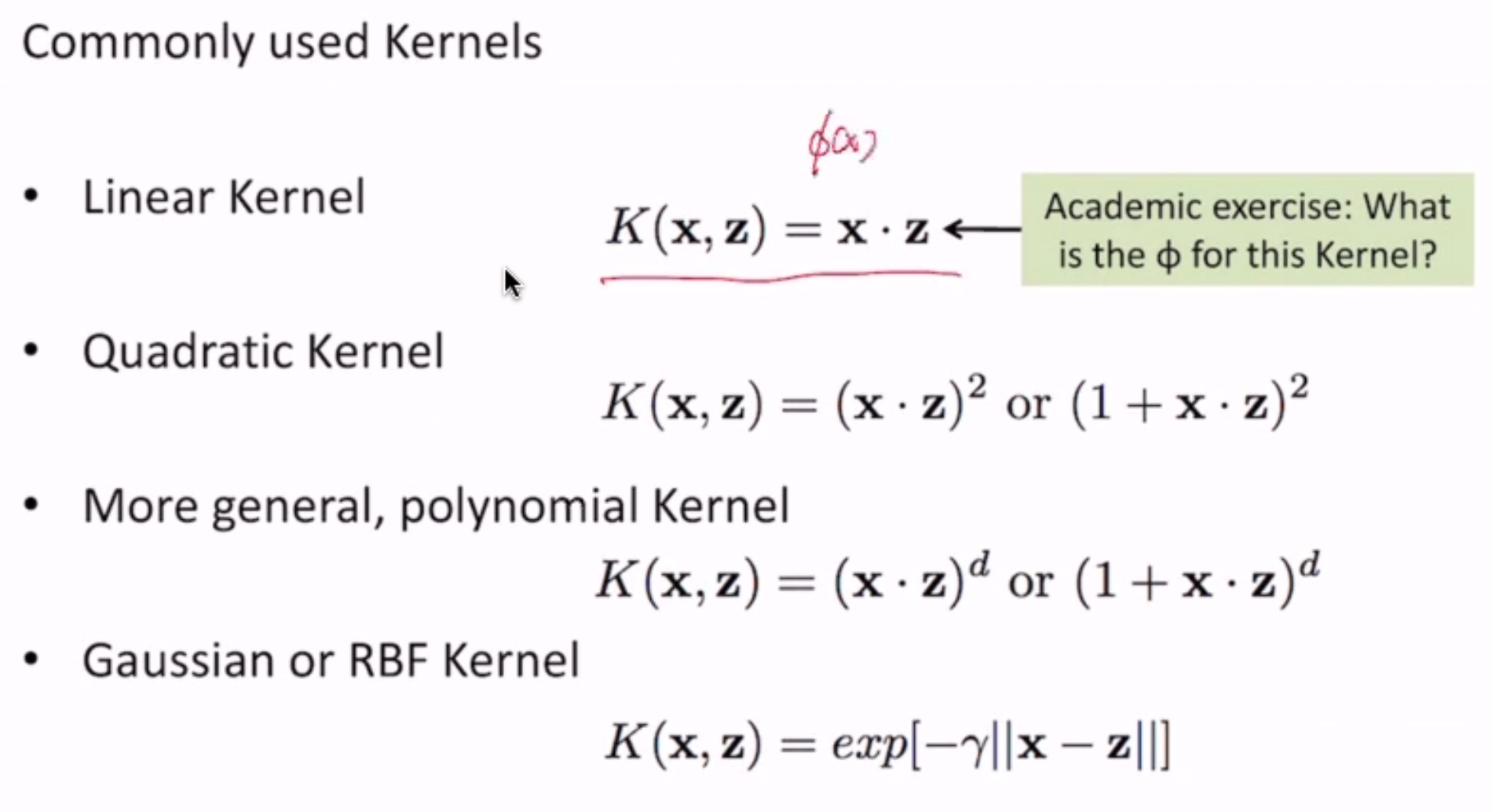
(\(\phi\) for the linear kernel = \(\phi(x) = x\))
Perceptron¶
we can also apply kernels to perceptron!
Naively, we can just replace \(\mathbf{x}\) with \(\phi(x)\) in the algorithm - but that requires knowledge of \(\phi\)

Prediction
since \(w = \sum_m \alpha_m \phi(x_m)\), prediction on \(x_n\) is easy: